The following illustrates the case of a small molecule mode-distortion and minimization.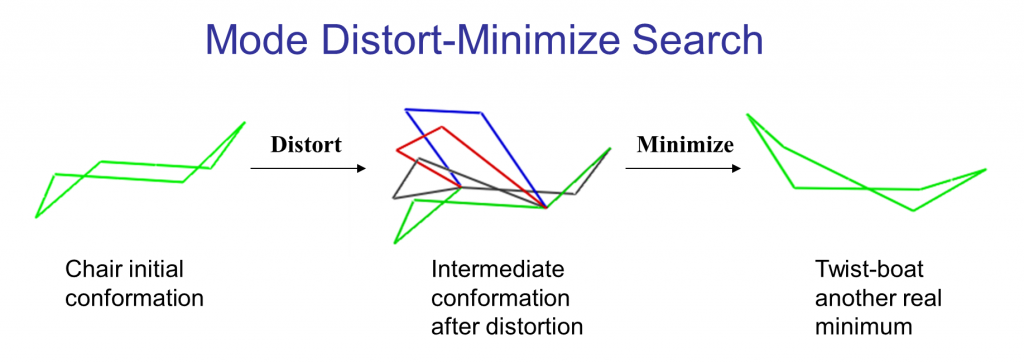 Each time a new lowest energy minimum is found it is subsequently used as a basis for the next mode distort-minimization. The number of torsional modes available depends on the size of the system.
Each time a new lowest energy minimum is found it is subsequently used as a basis for the next mode distort-minimization. The number of torsional modes available depends on the size of the system.
The search can cycle through the available modes in order (low to high energy) or at random. In addition, a linear combination of randomly chosen pairs of these modes can be used to define the distortion.
To further illustrate this approach consider the following six-atom system and its internal coordinates.
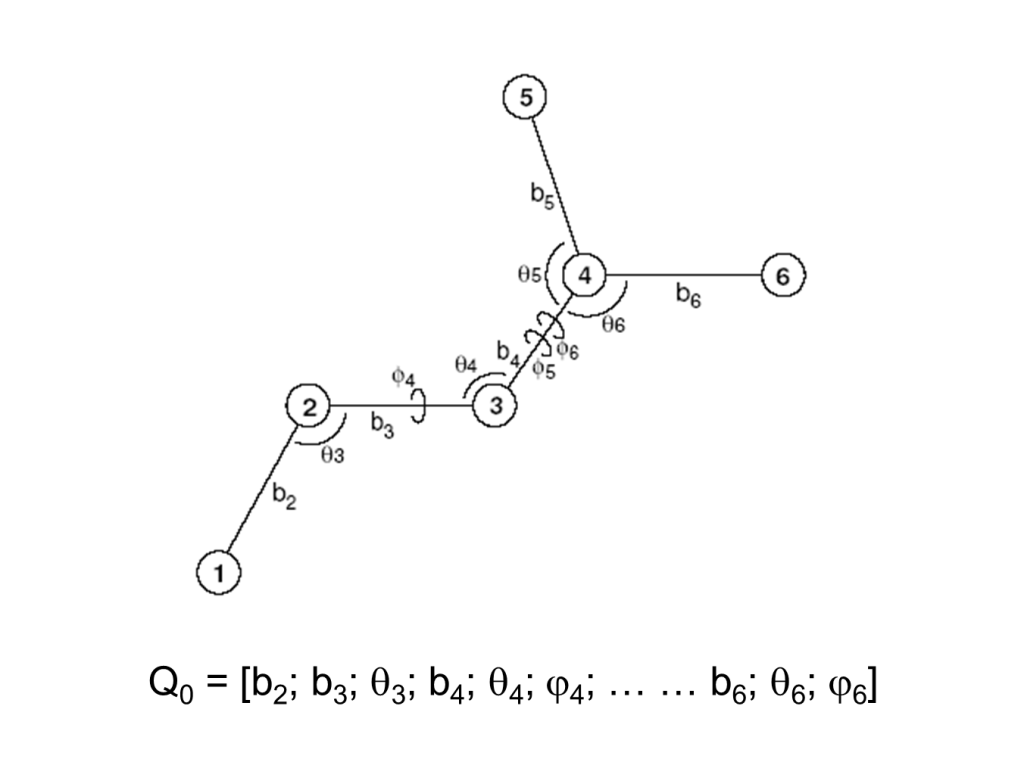 The total energy 2nd-derivative (Hessian) matrix with respect to internal coordinates (bond, angle, torsion coordinates) can be calculated
The total energy 2nd-derivative (Hessian) matrix with respect to internal coordinates (bond, angle, torsion coordinates) can be calculated