VCharge provides fast, easy access to accurate partial charges for virtually any drug-like compound. It is thus valuable in a wide range of modeling and QSAR applications. VCharge is available for Linux and Microsoft Windows®.
A convenient Windows viewer VDisplay is available for free download along with VCharge.
Key Strengths
Accuracy & Range
"Ab initio-like" partial atomic charges accurately reproduce electrostatic potentials from Hartree-Fock calculations at the 6-31G* level (SBKJC core potentials for iodine). See section on Accuracy, below.
Broad applicability
Verachem’s proprietary parameter set allows charges to be computed for virtually any stable compound composed of the elements hydrogen, carbon, oxygen, nitrogen, phosphorous, sulfur, fluorine, chlorine, bromine and iodine. Results do not depend upon molecular conformation.
Speed
Typically processes over twenty compounds per second on a modern desktop computer.
Convenience
Reads and writes SDfiles, and the free Vdisplay program allows graphical review of the results. Both Linux and Windows versions are available, and the Windows version of Vcharge includes a straightforward graphical user interface
Accuracy and Range of Applicability
VeraChem’s newest proprietary parameter set includes new atom types and has been thoroughly optimized. The new parameterization yields partial charges that accurately reproduce Hartree-Fock 6-31G* potentials at CHELPG sampling points. The potentials typically agree to within an RMSD of ~4 kcal/mol-e. For comparison, the previous parameters gave an RMSD of ~5 kcal/mol-e for the same set of over 350 molecules. Tests on molecules for which other charge sets have been published indicate that Vcharge is as accurate, relative to 6-31G* potentials, as much slower methods, such as RESP and AM1/BCC and is substantially more accurate than simpler approaches such as Gasteiger-Marselli.
Verachem’s new proprietary charges are strikingly similar to those in widely used force fields, such as AMBER and CHARMM22. Thus, for the 20 common amino acids, Vcharge matches CHARMM22 and AMBER94 with RMSDs of only 0.07 e and 0.10 e, respectively, and with correlation coefficients of 0.98 and 0.95. These differences are comparable to the differences between CHARMM22 and AMBER94 themselves: RMSD 0.10, correlation coefficient 0.90. Therefore, our new charges are compatible with these force fields and are appropriate for ligands when a protein is treated by the CHARMM22 or AMBER94 force fields.
How VCharge Works
VCharge is an electronegativity equalization method, where the electronegativity of each atom depends upon its atomic number, hybridization, and bonding environment within the molecule, and where special constraints are applied to keep too much charge from flowing off ionized groups. The constrained equalization problem is solved by an adaptation of the method of Lagrangian multipliers.
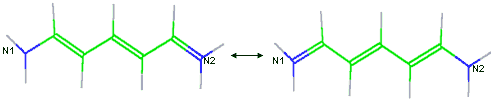
A unique feature of VCharge is its handling of alternate resonance forms. Alternate resonance forms of each molecule are automatically identified, and the atomic electronegativities and hardnesses are averaged over the resonance forms. This ensures that equal charges are assigned to chemically equivalent atoms, even when this equivalence is not apparent from the single resonance form provided in the input SDfile. For example, nitrogens N1 and N2 illustrated below would not be considered equivalent if resonance forms were not considered. Ionization and tautomer states are taken as specified in the input SDfile.
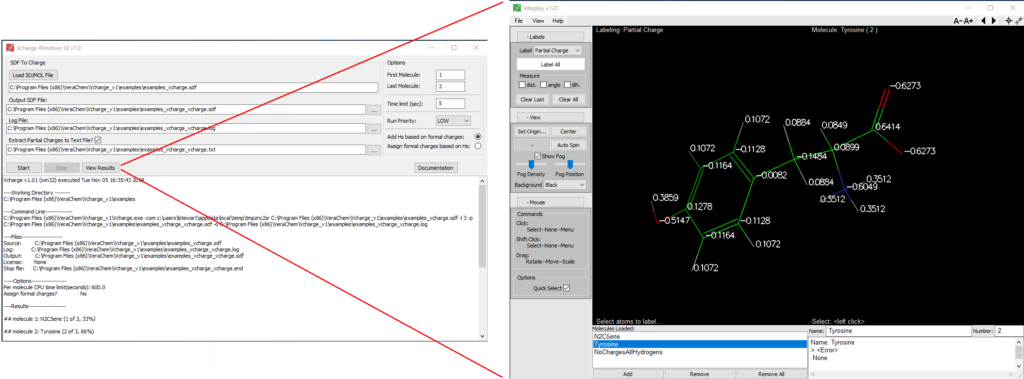
Windows User-Interface
Included with the Microsoft Windows® version of Vcharge, this convenient graphical user interface makes it particularly straightforward to run Vcharge and view its results. The interface brings together Vcharge, our molecular display program Vdisplay, and helpful Vcharge support utilities, described in the following section.
REFERENCES
Main Citations
Gilson, M. K., H. S. R. Gilson, and M. J. Potter (2003). “Fast assignment of accurate partial atomic charges: An electronegativity equalization method that accounts for alternate resonance forms.” J. Chem. Inf. Comput. Sci. 43: 1982-1997. DOI: 10.1021/ci034148o
Background Citations
Chen, W., J. Huang, and M. K. Gilson (2004). “Identification of symmetries in molecules and complexes.” Journal of Chemical Information and Computer Sciences 44(4): 1301-1313. DOI: 10.1021/ci049966a
Cornell, W., P. Cieplak, C. I. Bayly, I. R. Gould, K. M. Merz, D. M. Ferguson, D. C. Spellmeyer, T. Fox, J. W. Caldwell, P. A. Kollman (1995). “A 2nd generation force-field for the simulation of proteins, nucleic-acids, and organic-molecules.” Journal of the American Chemical Society 117(19): 5179-5197. DOI: 10.1021/ja00124a002
Bayly, C. I., P. Cieplak, W. D. Cornell, and P. A. Kollman (1993). “A well-behaved electrostatic potential based method using charge restraints for deriving atomic charges – the RESP model.” Journal of Physical Chemistry 97(40): 10269-10280. DOI: 0.1021/j100142a004
Breneman, C. M. and K. B. Wiberg (1990). “Determining atom-centered monopoles from molecular electrostatic potentials – the need for high sampling density in Formamide Conformational-Analysis.” Journal of Computational Chemistry 11(3): 361-373. DOI: 10.1002/jcc.540110311
Jakalian, A., B. L. Bush, D. B. Jack, and C. I. Bayly (2000). “Fast, efficient generation of high-quality atomic Charges. AM1-BCC model: I. Method.” Journal of Computational Chemistry 21(2): 132-146. DOI: 10.1002/(SICI)1096-987X(20000130)21:2<132::AID-JCC5>3.0.CO;2-P
Jakalian, A., D. B. Jack, and C. I. Bayly (2002). “Fast, efficient generation of high-quality atomic charges. AM1-BCC model: II. Parameterization and validation.” Journal of Computational Chemistry 23(16): 1623-1641. DOI: 10.1002/jcc.10128
Mackerell, A. D., J. Wiorkiewiczkuczera, and M. Karplus (1995). “An All-Atom Empirical Energy Function for the Simulation of Nucleic-Acids.” Journal of the American Chemical Society 117(48): 11946-11975. DOI: 10.1021/ja00153a017
Schmidt, M. W., K. K. Baldridge, J. A. Boatz, S. T. Elbert, M. S. Gordon, J. H. Jensen, S. Koseki, N. Matsunaga, K. A. Nguyen, S. J. Su, T. L. Windus, M. Dupuis, J. A. Montgomery (1993). “General Atomic and Molecular Electronic-Structure System.” Journal of Computational Chemistry 14(11): 1347-1363. DOI: 10.1002/jcc.540141112